Let pictures answer!
1 The first biomolecule simulated by computer
Question: what was simulated?
Left: X-ray structure of BPTI (bovine pancreatic trypsin inhibitor) molecule; right: structure after 3.2 ps of dynamic simulation (source: McCammon, Gelin, and Karplus, “Dynamics of Folded Proteins.”) This is the first biomolecular dynamics simulation. Molecular dynamics simulation simulates the status of molecular system during a certain time frame and gives out the change of system properties.
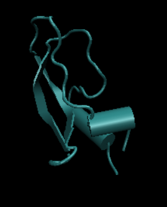 A model of BPTI molecule in VMD, a widely used program for visualizing molecules and the process of molecular dynamics simulation.
A model of BPTI molecule in VMD, a widely used program for visualizing molecules and the process of molecular dynamics simulation.
2 Creation of a scientifically meaningful environment of experiment in silico
Question: what were involved in the creation of the first “molecule in silico”?
Grounded on what?
-
Quantum mechanics’ success in describing some molecular structures and chemical reactions
The first five atomic orbitals. (Drawn using Orbital Viewer. Source: Wikimedia) Computer gradually became an indispensable tool for theoretical chemists after the World War II, and quantum chemistry was established as a sub-field in which computer simulation and calculation were widely used to obtain data that could not be got from real-world experiments. (Source: Gavroglu and Simões, Neither Physics nor Chemistry, 187.)
-
The early development in molecular dynamics simulation
This is a model of two water molecules (treated as two “rigid asymmetric rotors”) in minimum energy configuration. It was constructed for the first molecular dynamics simulation of a realistic system (i.e. a material system existed in the real world). (Source: Rahman and Stillinger, “Molecular Dynamics Study of Liquid Water.”)
By whom?
 Photo of Martin Karplus when he was awarded the Nobel Prize in Chemistry “for the development of multiscale models for complex chemical systems.” (Source: Wikimedia) The first biomolecular dynamic simulation was done by Karplus and his students, J. Andrew McCammon and Bruce R. Gelin. Both Karplus and McCammon had educational background in biology, physics and chemistry and desired to quantify molecular biology. This desire became a strong motivation for them to realize a computer simulation, which requires quantification in every step.
Photo of Martin Karplus when he was awarded the Nobel Prize in Chemistry “for the development of multiscale models for complex chemical systems.” (Source: Wikimedia) The first biomolecular dynamic simulation was done by Karplus and his students, J. Andrew McCammon and Bruce R. Gelin. Both Karplus and McCammon had educational background in biology, physics and chemistry and desired to quantify molecular biology. This desire became a strong motivation for them to realize a computer simulation, which requires quantification in every step.
How was it implemented?
Karplus and colleagues used computers in the Centre Européen de Calcul Atomique et Moléculaire (CECAM, European Center for Atomic and Molecular Calculations) to run the first biomolecular dynamics simulation. CECAM was an important academic center for early studies of molecular dynamics and other studies in theoretical chemistry.
 This is a picture of IBM 7094 in Columbia University in the 1960s. A computer of this model installed in mid-1960s’s MIT supported the first interactive 3-D modelling of biomolecules on screen. The development of computer technology would later enable more demanding tasks like Karplus and coworker’s work.
This is a picture of IBM 7094 in Columbia University in the 1960s. A computer of this model installed in mid-1960s’s MIT supported the first interactive 3-D modelling of biomolecules on screen. The development of computer technology would later enable more demanding tasks like Karplus and coworker’s work.
 The CDC 6600 and its successors were another series of widely deployed models for scientific computing in the 1960s and 70s.
The CDC 6600 and its successors were another series of widely deployed models for scientific computing in the 1960s and 70s.
Why was the simulation result regarded as valuable?
The early researchers did not emphasize that the simulated system in computer mirrored their real-world counterparts or use the visual similarity and impact to argue for the new method’s legitimacy. Biomolecular dynamics simulation was accepted by the Karplus group and other scientists as a legitimate scientific tool because it provides information that experimental methods could not give.
3 After the first biomolecule in silico: simulation software toolkit became digital scientific instrument
Question: why has molecular dynamics simulation become more and more popular?
An example of materialization and encapsulation of knowledge into a “toolkit”, the CHARMM program. (Source: https://www.charmm.org/) With such well-packed program, scientists can do simulation without necessarily being good at programming. The credibility and potential value was supported by previous studies that using the same software toolkit.
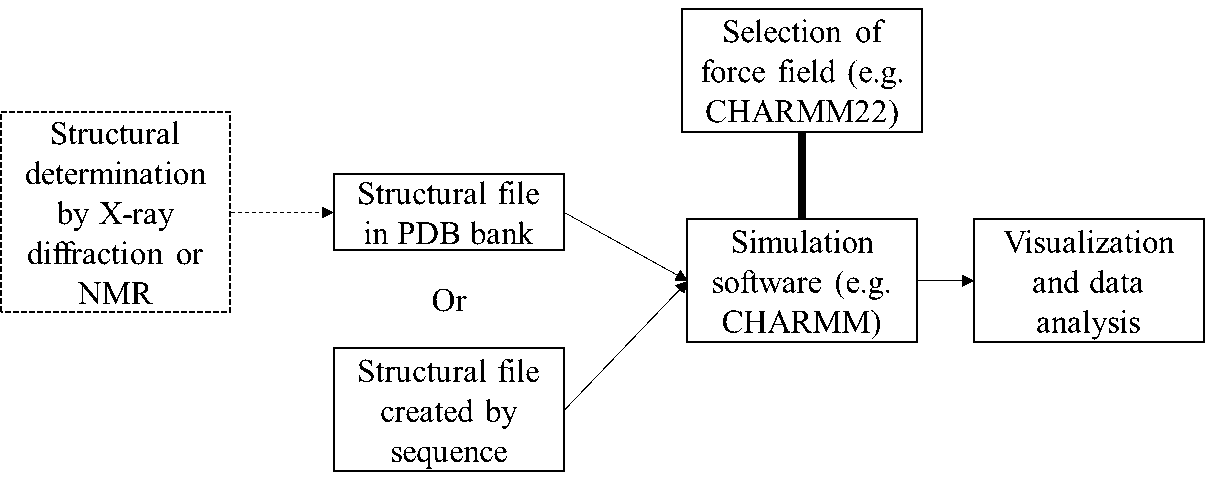 A typical workflow of molecular dynamics simulation. This workflow has been standardized, just like the workflow of implementing other kinds of analytical methods (e.g. NMR, GC-MS).
A typical workflow of molecular dynamics simulation. This workflow has been standardized, just like the workflow of implementing other kinds of analytical methods (e.g. NMR, GC-MS).
Bibiography
Gavroglu, Kōstas, and Ana Simões. Neither Physics nor Chemistry: A History of Quantum Chemistry. Cambridge, MA: MIT Press, 2012.
Leach, Andrew R. Molecular Modelling: Principles and Applications. 2nd ed. Harlow, England ; New York: Prentice Hall, 2001.
Levinthal, Cyrus. “Molecular Model-Building by Computer.” Scientific American 214, no. 6 (1966): 42–53.
McCammon, J Andrew, and David Allison. “J. Andrew McCammon Interview,” 1995. https://mccammon.ucsd.edu/people/pi/interview.php.
“CHARMM.” Accessed September 9, 2018. https://www.charmm.org/.
Rahman, Aneesur, and Frank H. Stillinger. “Molecular Dynamics Study of Liquid Water.” The Journal of Chemical Physics 55, no. 7 (1971): 3336–59.
Comments